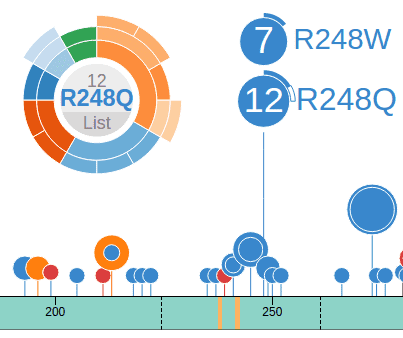
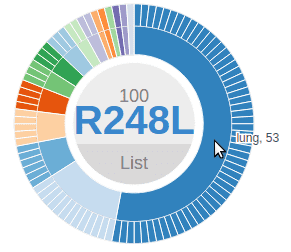
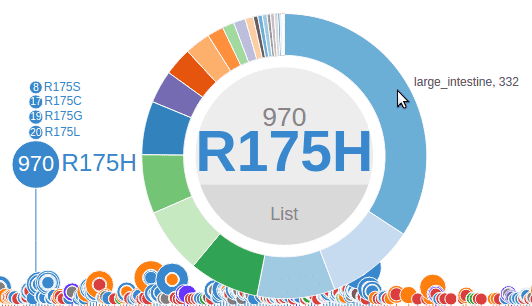
In the Pediatric data set, clicking on a disc will bring up a sunburst chart showing the cancer type and subtype information for the set of samples affected by this mutation. The sunburst chart is labeled by amino acid change in the center and can be moved by clicking and dragging on this label:
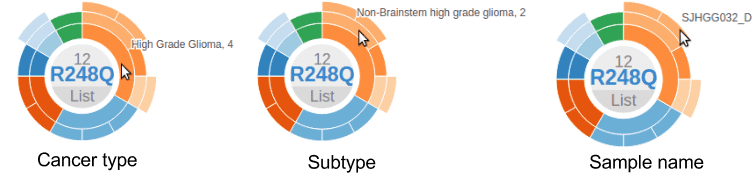
In a sunburst chart, the inner layer represents cancer types. The outmost layer represents each individual sample. When there is cancer subtype information available for the sample, an intermediate layer will appear. Hovering cursor on any layer will display the name:
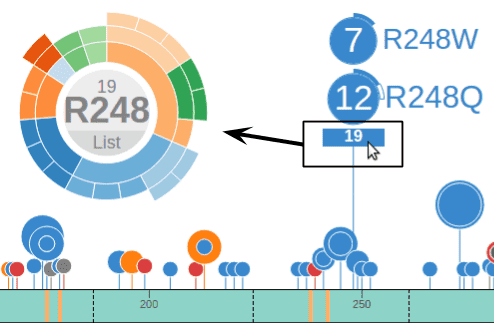
Mousing over the bottom of the R248Q disc will show a box with number 16, representing all 16 mutations at the position R248 (11 from R248Q, 5 from R248W). Clicking this box will bring up a new chart representing these 16 mutations. Notice the label at the center now reads “R248”, indicating it now represents all amino acid changes at R248:
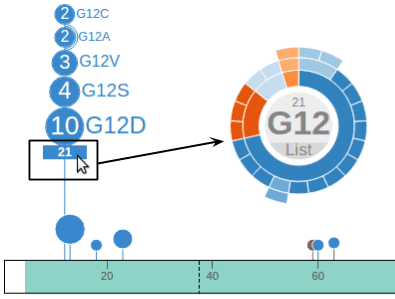
This feature is handy for exploring cases like the KRAS mutations at G12, a hotspot with multiple mutated alleles. A single click will show the cancer type composition for all different types of G12 mutations:
For COSMIC, clicking on a disc will also generate a sunburst chart but consisting of only two layers: the inner layer representing the source tissue type of the cancer, and the outer layer representing samples:
As an optimization to improve performance, when the number of mutations is greater than 400, the sunburst chart will not show individual samples at the outmost ring:
To dismiss the chart, click on the center of the ring around the mutation name.
Click the “List” button inside the chart to show a mutation table, as described below.
Using the mutation table
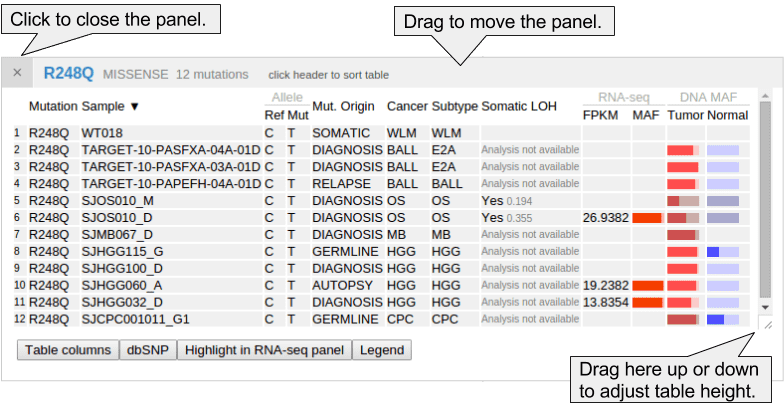
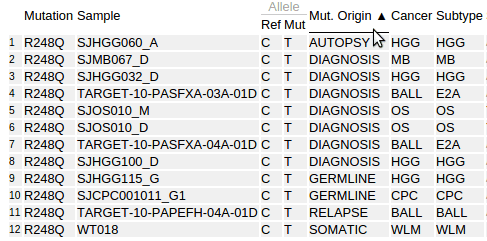
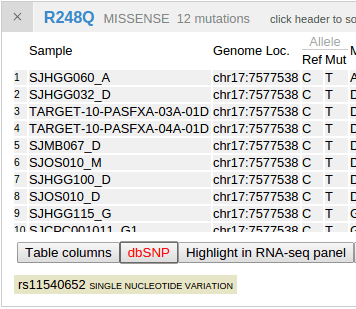
The mutation table is a powerful and versatile tool for examining details about samples and mutations. The following table shows details for the pediatric cases of TP53 p.R248Q:
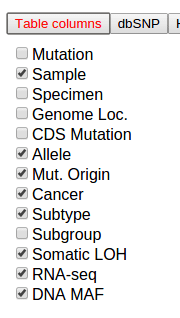
Additional columns that are not shown by default may be added by clicking the button “Table columns”, which shows a list of all available columns. Toggle the checkboxes to add/remove columns to the mutation table. The following shows the column listing for the Pediatric dataset:
Clicking on the header of any column in the table will sort the table based on the values in that column. Sorted columns will show a black up/down arrow in their header. Clicking the header again will reverse the sorting order:
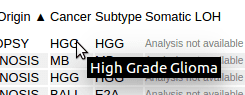
In the Pediatric mutation table, the cancer types are shown in abbreviated forms. Hover the cursor over the abbreviations to display their full names:
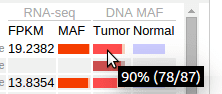
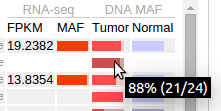
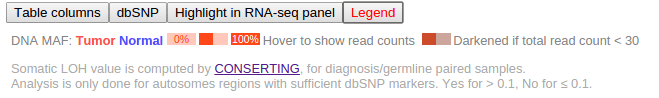
In Pediatric data set, mutant allele fraction (MAF) information is available for the DNA of tumor and normal genomes, and for tumor RNA-Seq of selected samples. This information is displayed in graphical forms of “percentage bars”. Hover over a bar to see the fraction value and read counts:
If the total read count is below the cutoff of 30, the bar will appear darkened:
MAF value graphical display is also explained in the table legend, brought up by clicking the “Legend” button:
Loss of heterozygosity (LOH) information will be shown in the mutation table for part of the Pediatric samples, which is also explained in the legend above.
dbSNPs that are overlapping with mutations will be identified and shown along with the mutation table. Click the second button “dbSNP” in the bottom to show any such hits. Click on the displayed SNP to view its record in the dbSNP database:
The button labeled “highlight in RNA-seq panel” is explained in section [put link here].
Zooming in to browse mutations at nucleotide level
To Do:
We are still working on importing this content to the official St. Jude Cloud guide. Please be patient with us! If you have any questions, please contact us.
Original Link: https://plus.google.com/u/0/+XinZhou_s/posts/5Ji9ZkecaDD