Using heatmap
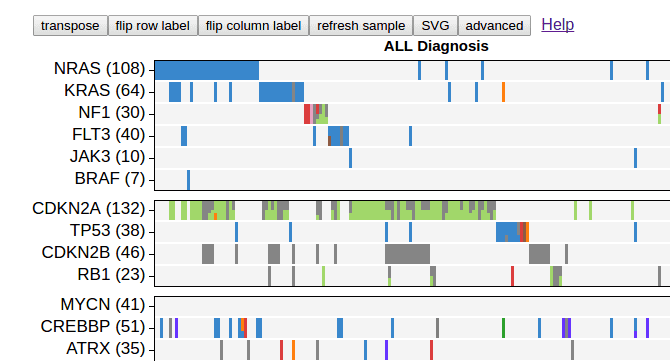
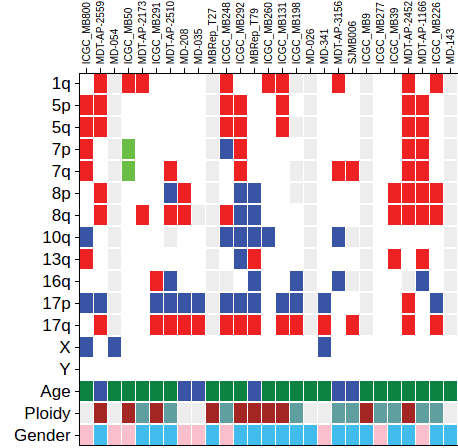
Heatmap is a gene-by-sample grid representing mutations from a data set, looking like below:
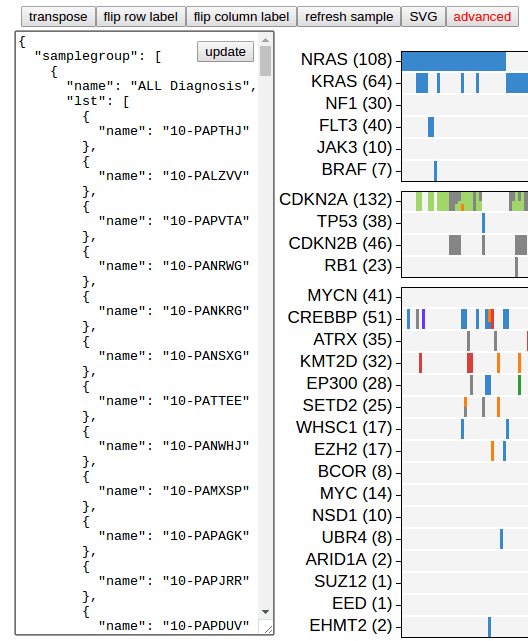
The heatmap can be customized to great extent. Click the button “advanced” on top:
A box opens on the left showing texts that can be edited. The content is an JSON object describing the heatmap. Make modification to the JSON content, click “update” button to render a new heatmap to reflect your changes.
“Save your map before leaving…”
ProteinPaint will not save the JSON when you leave, because the heatmap is based on user-provided data. If you have made edits, copy and save the JSON text before leaving ProteinPaint. Next time you explore the same dataset, launch the heatmap, and paste your saved JSON in the text box to re-create the heatmap.
“Show a preconfigured map”
- A preconfigured map can be stored in the schematics of a study view. Refer to the ”heatmapJSON” attribute of the study schematics
- Also in study view, try the ”hard-coded map” in which you have complete control of what is showing in one or more maps.
- To embed a map in your website, use the “studyview” as instructed here.
Genes
Sort samples by number of mutation in selected genes
{"name": "JAK3", "sortorder": 1 }By applying the “sortorder” attribute to a gene, samples will be sorted by the descending order of number of mutations in this gene. Samples with more mutation in this gene will be moved to left (or top).
Value is any number.
Multiple genes can be used for sorting. In such case, the numerical value determines the precedence of which genes are taken into sorting (smaller the value, earlier the gene is used).
Samples are sorted within each sample group.
Default applied to all genes.
To disable such sorting, remove the attribute from a gene.
Gene order
The order of genes is determined by the order of appearance of genes in each gene group in the heatmap JSON.
[ {"name": "JAK2"}, {"name":"PAX5"} ]In this example, JAK2 appears earlier than PAX5.
By default, genes are arranged in descending order of total number of mutations from all samples.
Samples
Sample order
By default, the order of samples are automatically adjusted using the “sort-by-gene” (described above).
To manually adjust sample order, the sorting-by-gene must be disabled first. Then edit the order of samples in the sample group section of the heatmap JSON to take effect.
Sample group
Samples can be arranged into groups. Space can be applied between groups and borders can be drawn around each group, so that they appear distinct visually.
{"samplegroup": [
{
"name":"sample group 1",
"lst":[
{ "name":"sample_1" },
{ "name":"sample_2"}
]
},
{
"name":"sample group 2",
"lst":[
{ "name":"sample_3" },
{ "name":"sample_4"}
]
}
]
}This code defines two sample groups, each with two samples.
The “samplegroup” attribute is root-level.
Refresh sample
You can do this by clicking the “refresh sample” button located on top, rather than editing the JSON. This is because by default the heatmap chooses samples by requiring the sample to have at least 1 mutation in the initial set of genes. Afterwards the set of samples stay unchanged unless you alter the “samplegroup” contents in JSON.
Sometime when user started using a new gene set different from the initial set, he/she would like the samples to update accordingly. This is when the “refresh sample” button comes in handy.
Be careful that refreshing samples and your previous edits on samples will be lost.
Will be available in the next release.
Border
Sample/gene group border
{"border":"black",
"borderwidth":1}Add border attribute to either a sample or gene group, and a solid border in specified color will be drawn enclosing this group.
Optionally provide “borderwidth” attribute to specify the width using integer value. Default 1.
Default applied to gene group.
Color
Heatmap cell background color
{"cellbg":"#f1f1f1"}The background fill color for a heatmap cell if there is no data. Applicable for both data cells and metadata cells. Root-level attribute. Default provided.
Sample group background color
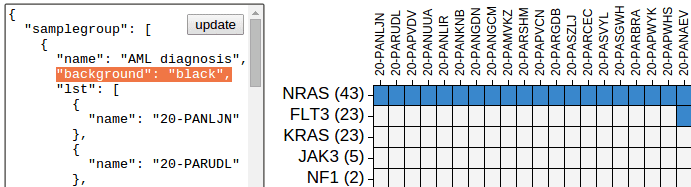
{"background":"black"}Add background attribute to a sample group. The entire group will show this background color. Default not applied.
This can be used to generate the look of “borders” around heatmap cells. The border width will be controlled by row/column spacing.
Cell background color by sample
{"name":"20-PANLJN", "cellbg":"#CCCCCC"}Add the cellbg attribute to a sample. All cells belonging to this sample will use the specified background color instead of the global background color. Default not provided.
Will be available soon.
"metadata":[]
Metadata is used for annotating samples.
This is deprecated. Try to use ”metadatabymatrix” instead.
Example
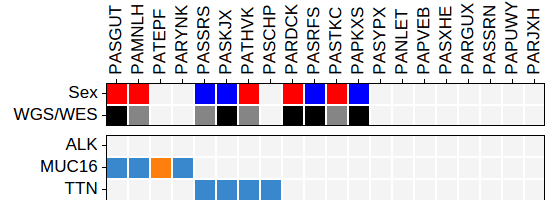
"metadata":[
{"key":"a",
"label":"Sex",
"type":"fill",
"values":{
"male":"red",
"female":"blue"
}
},
{"key":"b",
"label":"WGS/WES",
"type":"fill",
"values":{
"WGS":"black",
"WES":"#858585"
}
}
],
"patientannotation":{
"PARDCK":{"a":"male", "b":"WGS"},
"PASRFS":{"a":"female","b":"WGS"},
"PASTKC":{"a":"male", "b":"WES"},
"PASSRS":{"a":"female","b":"WES"},
"PASGUT":{"a":"male", "b":"WGS"},
"PASKJX":{"a":"female","b":"WGS"},
"PATHVK":{"a":"male", "b":"WES"},
"PAPKXS":{"a":"female","b":"WGS"},
"PAMNLH":{"a":"male", "b":"WES"}
},
"metadataborder":"black",Rendered
Metadata terms
"metadata":[ ... ]Root-level attribute.
Value is a list, with each metadata term as one element. A term is defined as an object like below:
{
"key":"term1key",
"label":"Term name",
"type":"fill",
"values":{
"term1attribute1":"red",
"term1attribute2":"blue"
}
},key: key identifier of this term
label: name to be displayed
type: “fill” only, reserved for future expansion
values: attribute values for this term. Key is attribute name, value is rendering color.
Metadata box border and color
"metadataborder":"color",Root-level attribute.
If present, draw border around metadata section with given color.
Width of metadata box border
"metadataborderwidth":2,Root-level attribute.
If present, draw border with line of given thickness.
"patientannotation":{ }
Root-level attribute.
For annotating patients or individuals.
To be used along with the “metadata” attribute
Value is a hash, with patient names as keys, and annotations as values.
{
"sample1":{
"term1key":"term1attribute1",
"term2key":"term2attribute1",
},
"sample2":{ ... },
...
},When there is no sample type in the dataset, must use this attribute but not “sampleannotation”.
When there IS sampletype, using this attribute will annotate patients, but not the separate samples. To annotate samples instead (e.g. diagnosis/relapse), use “sampleannotation” described below.
"sampleannotation":{ }
Root-level attribute.
For annotating individual samples rather than patients.
To be used along with the “metadata” attribute.
Value is a nested hash, with patient/individual name as level-1 keys, sample type as level-2 keys, and sample annotations as values.
{
"patient1name":{
"sampletype1":{
"term1key":"term1attribute1",
"term2key":"term2attribute1"
},
"Sampletype2":{
... annotation to this sample in this patient
}
},
"patient2name":{ ... },
...
},"metadatabymatrix":{}
Work in progress!!
Prepare metadata annotation in a spreadsheet and declare it using ”metadatabymatrix“.
Example:
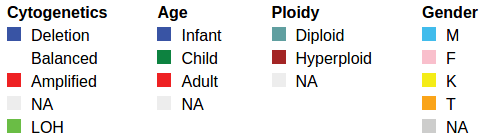
"metadatabymatrix":{
"annotations":[
{"text":"matrix data lines"}
],
"terms":{
"Cytogenetics":{
"Deletion":{ "label":"Deletion", "color":"#3954a4" },
"Balanced":{ "label":"Balanced", "color":"white" },
"Amplified":{ "label":"Amplified", "color":"#ee2123" },
"NA":{ "label":"NA", "color":"#ededed" },
"LOH":{ "label":"LOH", "color":"#6abd45" }
},
"Age":{
"infant":{ "label":"Infant", "color":"#3954a4" },
"child":{ "label":"Child", "color":"#0d8240" },
"adult":{ "label":"Adult", "color":"#ee2123" },
"NA":{ "label":"NA", "color":"#ededed" }
},
"Ploidy":{
"diploid":{ "label":"Diploid", "color":"#609fa0" },
"hyperploid":{ "label":"Hyperploid", "color":"#a32525" },
"NA":{ "label":"NA", "color":"#ededed" }
},
"Gender":{
"M":{ "label":"M", "color":"#41bcec" },
"F":{ "label":"F", "color":"#f9bfcc" },
"K":{ "label":"K", "color":"#f4ec17" },
"T":{ "label":"T", "color":"#faa419" },
"NA":{ "label":"NA", "color":"#ccc" }
}
}
}Metadata rows “1q” to “Y” belongs to the group “Cytogenetics”, as defined in metadatabymatrix.terms:{}.
In the legend, it will show “Cytogenetics” but rather each chromosome arm like 1q, 5p and such.
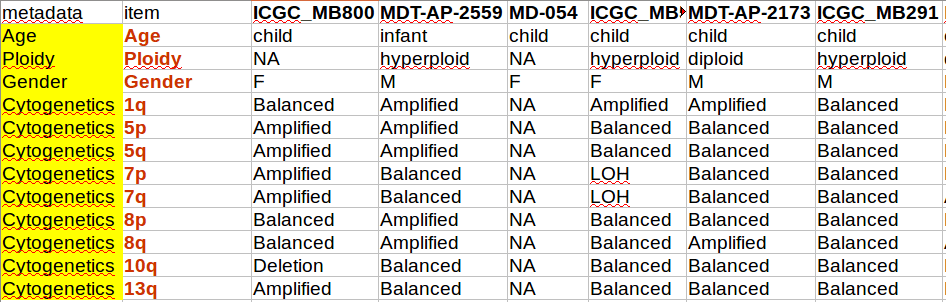
Example of a matrix viewed in spreadsheet:
Content of this file will be represented as a single string, where columns are joined by “\t” and lines joined by “\n”.
Others
Gene recurrence
"samplecount4gene":true Root-level attribute.
If set to “true”, gene recurrence will be indicated by the number of samples: