Pediatric and COSMIC are the two data sets formally supported by ProteinPaint. These are indicated by two buttons on the top, referred to as handles. Click the handle labeled “Pediatric” to load Pediatric mutations:
Once loaded, the Pediatric mutations will be shown as discs extending from the protein, and the handle will turn gray, indicating the data has been loaded. The top of the graph shows a header with a set of buttons starting with “Pediatric”:
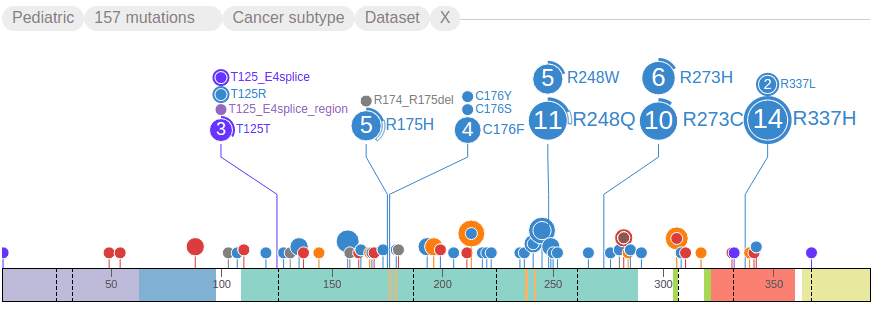
Click the handle for “COSMIC” to load adult cancer mutations. This can take a few moments to load since the amount of COSMIC mutations may be much larger than Pediatric, depending on specific protein. In the following example, COSMIC data is shown on the other side of the protein from the Pediatric data set, allowing for convenient comparison. On the COSMIC side of the graph, discs extend downward from the protein, and the header is at the bottom:
At this stage, you may click either “Pediatric” or “COSMIC” handle to hide the graph from ProteinPaint display, after which the handle turns dark gray again:
Click on the handle to toggle the the data set on again.
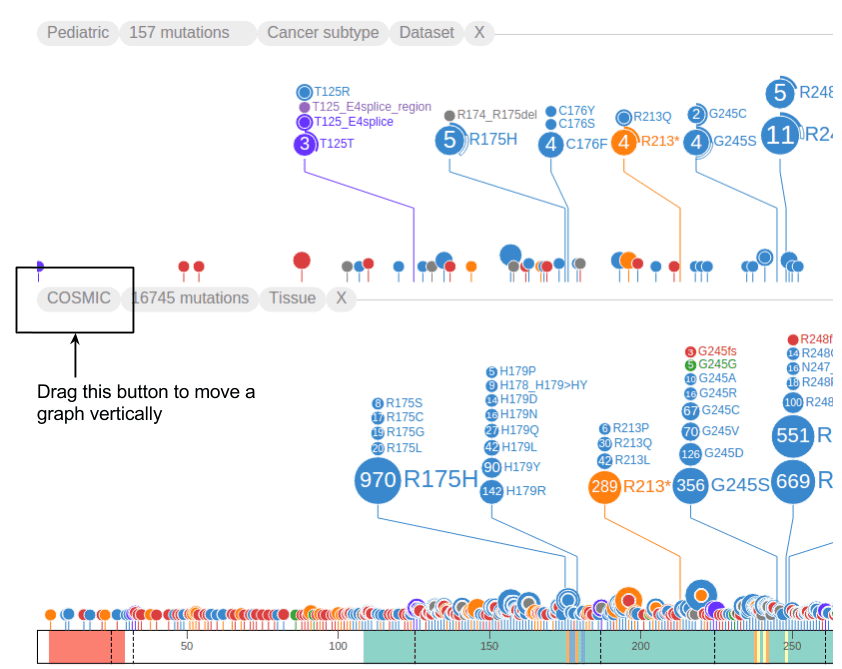
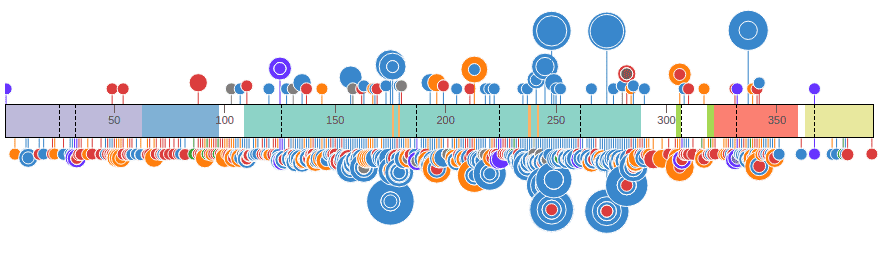
Dragging the first button in the graph header moves the graph vertically. It can be moved to the top or bottom of the protein ruler, or change its order of appearance when there are multiple data set graphs. The following shows both COSMIC and Pediatric data sets on top of the protein:
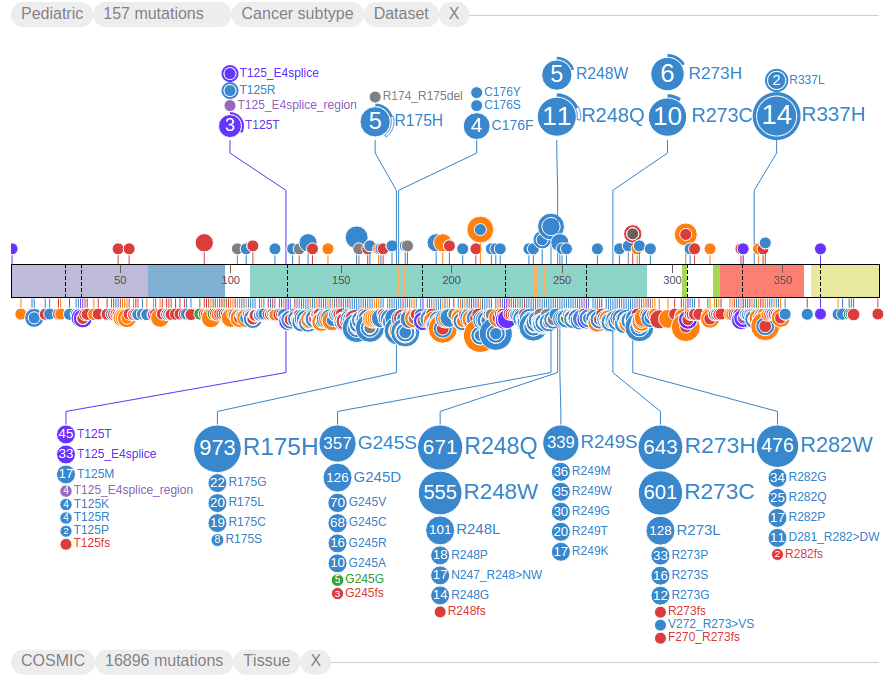
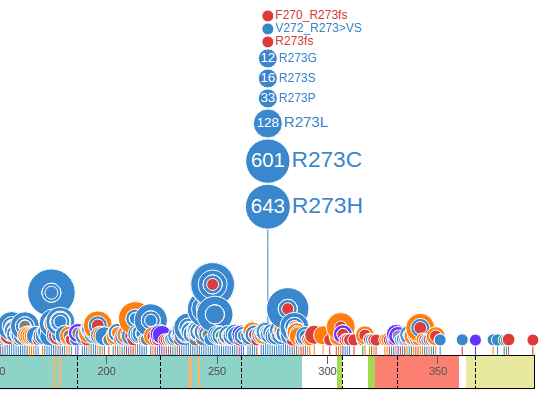
ProteinPaint shows the full view of the protein by default, and groups mutations by amino acid positions, so that discs representing mutations at the same amino acid position will be aligned to the same horizontal position. At each amino acid position, discs are vertically stacked by the number of mutations in each group, with largest group closest to the protein, as illustrated by an example from COSMIC:
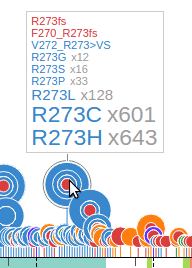
The text labels to the right of the discs show amino acid changes. Together the disc radii and label font sizes reflect the number of affected samples by each mutation. Clicking on any text label will fold the discs and their labels. Clicking on a disc will expand its folded mutation. These interactive features allow the user to configure and customize the fold/expand states of any mutation as desired. For added convenience, an option is provided allowing the user to toggle the fold/expand of all mutations in a dataset by clicking on the second button of the graph header. The following shows all TP53 mutations folded using this method:
When folded, the discs will be slightly raised away from the protein by a distance relative to the number of mutations at that amino acid position. To get a quick view of the data contained in any folded disc, mouse over the disc and a tooltip will appear, for example:
ProteinPaint highlights mutations of germline and relapse origin using arcs surrounding the discs. In the example below, the 11 mutations for R248Q show two types of arcs, the solid arc indicates two of these mutations are from germline samples, and hollow arc indicates one is from a relapsed sample. The remaining portion of the disc not covered by an arc indicates the final 8 mutations are from diagnostic (somatic) samples:
At the present time, only the Pediatric dataset contains germline and relapse mutations. All COSMIC mutations are somatic.