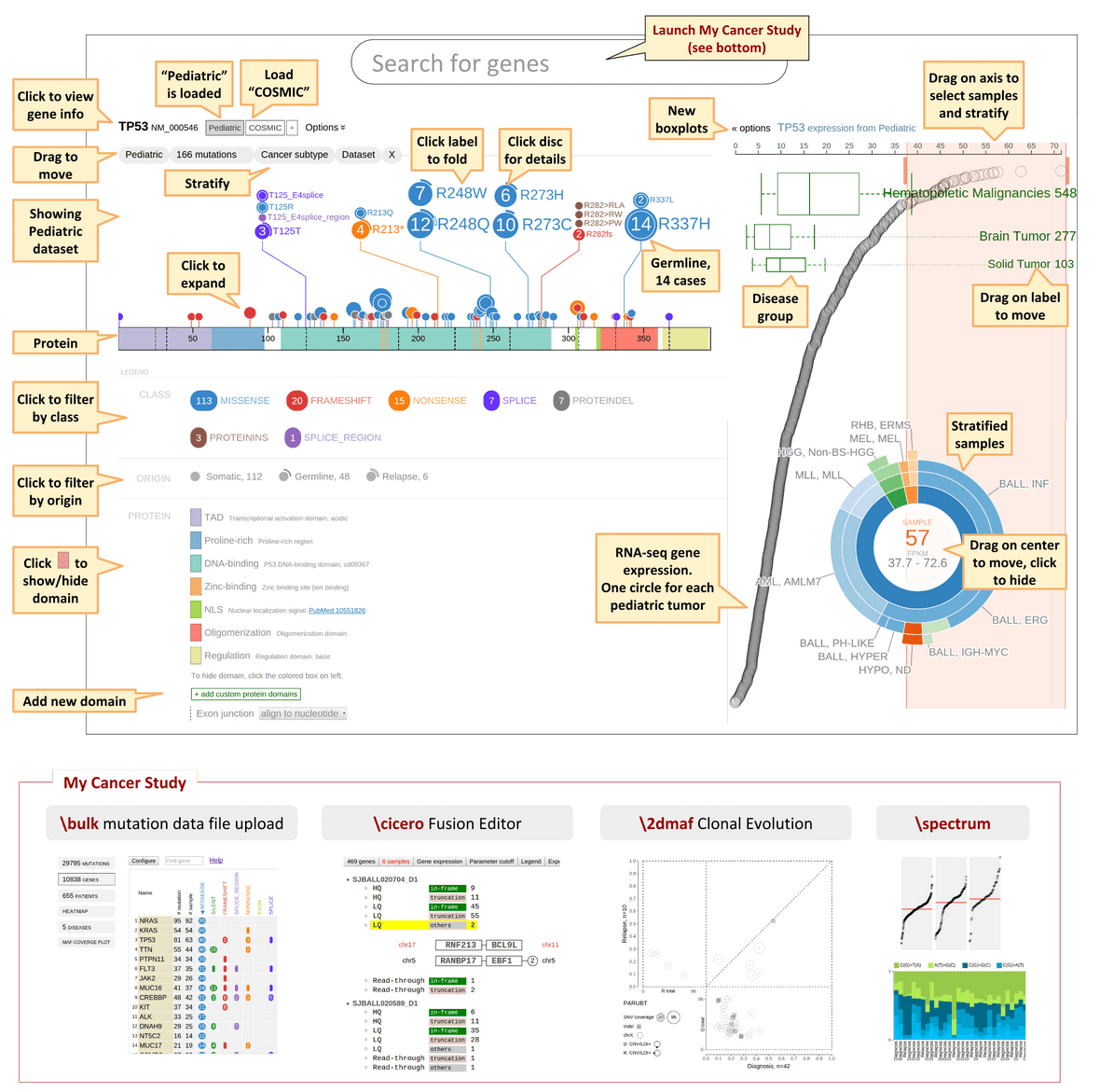
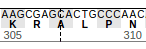Use the search box on the top to find a protein by HUGO gene symbol, for example “TP53”.
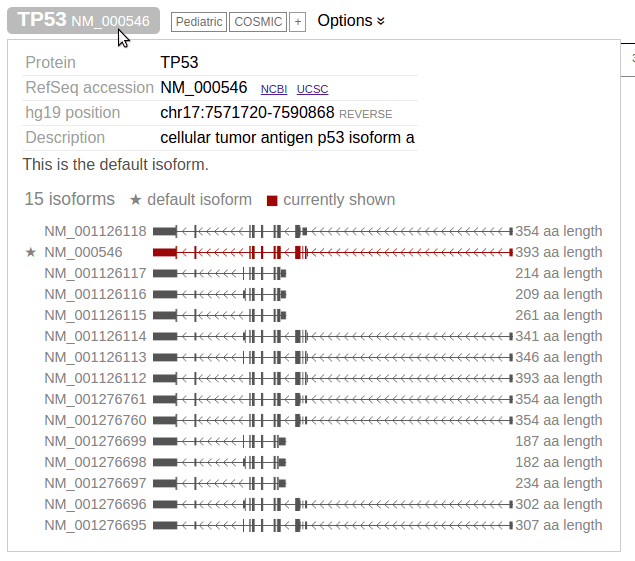
The TP53 protein is rendered as a color-banded ruler, with the numerical axis the representing number of amino acid residues from N-terminus to C-terminus. This example shows the protein translated from the preferred isoform of TP53 gene (NM_000546), shown on the top left. Click the gene symbol to view all isoforms for TP53:
To view a different protein isoform, either click on the diagram of the isoform in above panel, or search by the isoform’s RefSeq accession number instead of gene symbol.
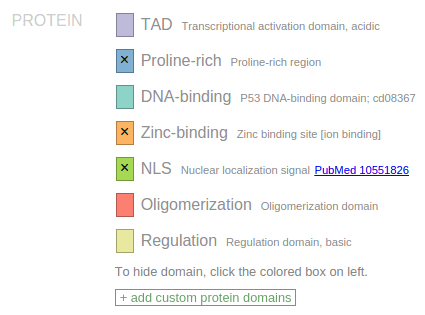
Protein domain
The source of protein domain annotation is primarily the NCBI CDD database, which has been supplemented by in-house curation efforts. In ProteinPaint, domains are represented by color bands, with varying colors indicating different types of domains. The list of domain types is shown in the legend according to their order of appearance on the protein. For manually curated domains, a PubMed link may be available to reference the source publication.
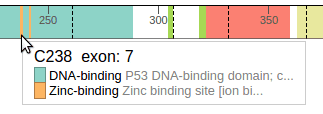
Hovering the cursor over the protein will display a tooltip showing any domains under the cursor (in some cases there can be more than one):
As a convenience, ProteinPaint offers an option to hide selected domains. To hide a domain type, click on one of the colored boxes in the legend to hide all occurrences of the selected type of domain from display. An “x” sign will appear inside the box to indicate a domain has been hidden. Following example shows TP53 protein with three types of domain hidden:
Hidden domains will be excluded from screenshots. Click the box marked by “X” to toggle the domain visible again.
Exon junction
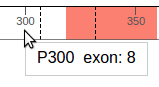
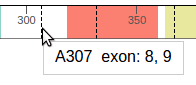
When hovering the cursor over the protein, the tooltip indicates the amino acid residue under the cursor, and the associated exon number:
Dashed lines represent exon junctions. The following example shows an exon junction is overlaps with Alanine 307, which is marked by exon numbers 8 and 9:
To show additional details about A307, click on this position and ProteinPaint will zoom in to show that the exon junction falls inside codon A307, between the first and second nucleotides. The codon nucleotides are shown in the top row, and amino acid residues are shown in the middle. Alternating background shades help distinguish adjacent codons. During zoom-in, the genomic coordinates of nucleotides will also be shown in tooltip:
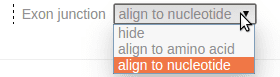
The exon junctions are aligned to nucleotide positions by default. This alignment can be configured or turned to hidden using a drop-down menu in the legend:
For example, by choosing the option “align to amino acid”, the exon junction inside A307 will be aligned to the center of the A307 residue:
After zooming-in, clicking on either side of the protein will scroll left or right. To zoom out and view entire protein again, click the “Show all” button on the top: