Fusion gene data are available for a subset of Pediatric cancer samples (Ph-like B-cell acute lymphoblastic leukemia, Fig. 1b), detected from RNA-Seq data using the software CICERO (Li, Y. and Zhang, J. in preparation). The comprehensive set of fusion gene information from all Pediatric samples is being produced at the time of writing and will be made available on ProteinPaint.
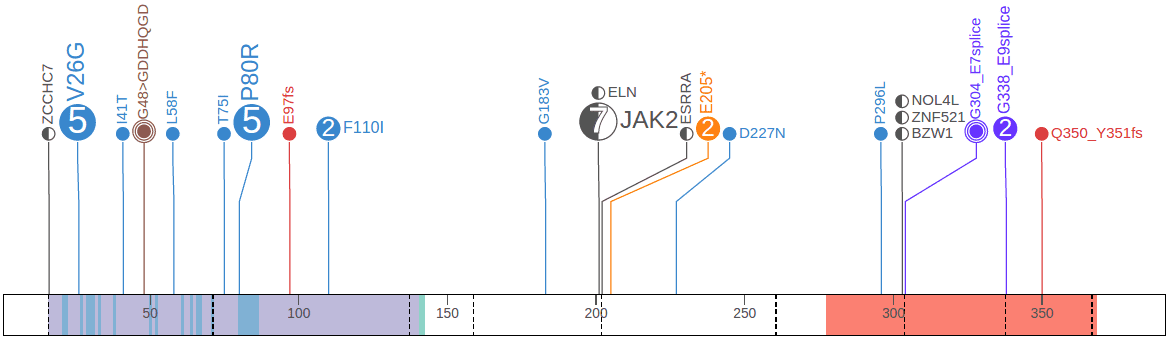
As an example, viewing Pediatric mutations for PAX5 shows a number of fusion gene events amongst SNVs and indels:
In this graph, there are 7 cases of fusion with JAK2. The disc is half-filled on left indicating the N-terminus part of PAX5 protein is fused with JAK2.
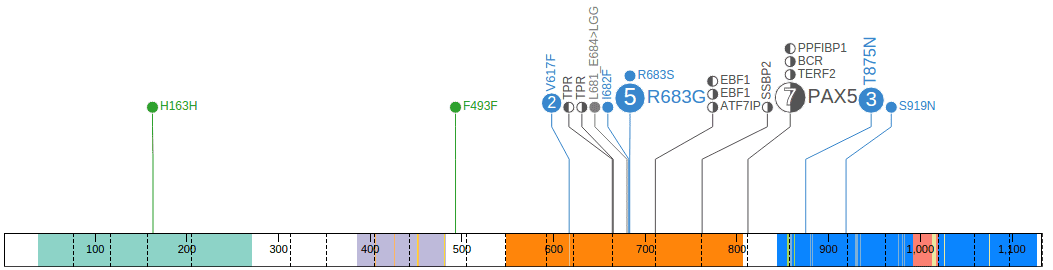
Conversely, in JAK2 ProteinPaint shows the same set of 7 fusion events with PAX5:
Click on the half-filled disc labeled “PAX5” to view a graphical representation of this fusion event:
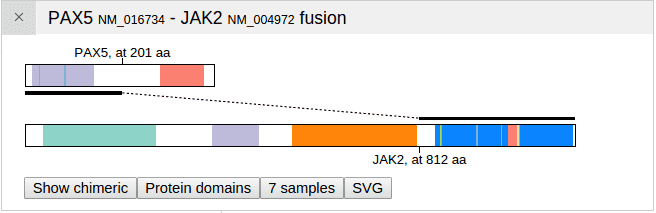
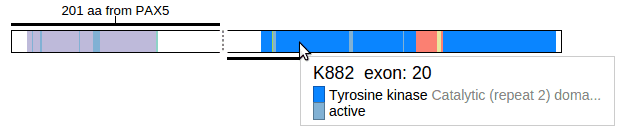
In this view, the protein schema is displayed for both PAX5 and JAK2. The two proteins are positioned in this way that the N-terminus segment (PAX5) is on top, and the C-terminus (JAK2) is on bottom. A thick horizontal line marks out the part of each protein that is retained in the fusion event, and a thin dashed line joins the segments. The amino acid positions from which the dashed line starts and ends are marked on both proteins.
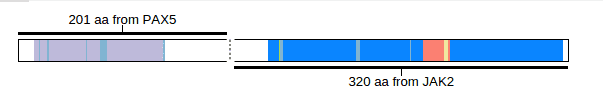
Click the button “Show chimeric” to transform the view into a “chimeric” protein, in which only the N-terminus of PAX5 and C-terminus of JAK2 are visible:
Mouse over either of the proteins to examine details about protein domains through the tooltip:
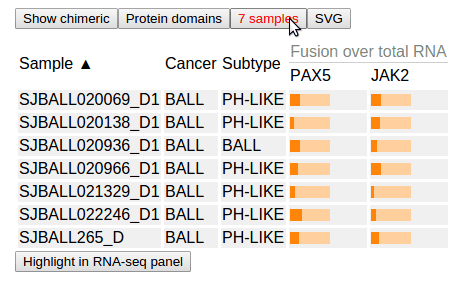
Click the button “7 samples” to view the list of samples from which the PAX5-JAK2 fusion has been discovered:
In this table, “Fusion over total RNA” shows color-bars to show the fraction value of fusion transcripts in the total transcripts of each gene (PAX5 and JAK2). This fraction value is calculated by the software CICERO.