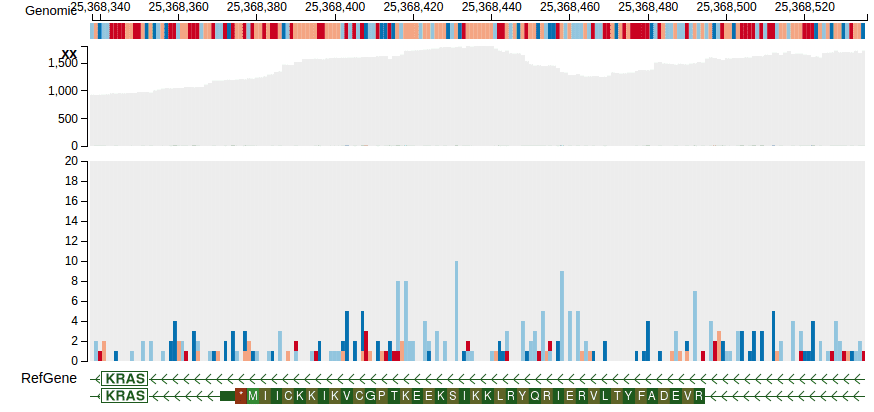
“Bampile” track
File format
The file has 3 columns:
- Chromosome name
- Basepair position, 0-based
-
JSON object, e.g.
- {“Raw0”:{“A”:8,“C”:11,“G”:5,“T”:4050},“Raw30”:{“A”:4,“C”:7,“G”:4,“T”:3966},“Raw35”:{“A”:3,“C”:7,“G”:3,“T”:3771},“Raw38”:{“A”:1,“C”:5,“G”:3,“T”:3334},“New0”:{“C”:4,“T”:2652},“New30”:{“C”:4,“T”:2633},“New35”:{“C”:4,“T”:2598},“New38”:{“C”:2,“T”:2382}}
- Keys are various grades
- For each grade, read depth for each observed nucleotide is given in an object
Prepare a track
To prepare bampile track file from Xiaotu’s output in St. Jude internal system:
Node.js v6.0 and above is required. Linux binaries are available at https://nodejs.org/dist/v6.9.1/node-v6.9.1-linux-x64.tar.xz
(Do not load Node.js using “module load”, that’s outdated)
module load tabix
cd /research/dept/compbio/common/proteinpaint-dev/tp/ultra
node bampileparse.js SAMPLE.input.txt > tempfile
sort -k1,1 -k2,2n tempfile > SAMPLE
bgzip SAMPLE
tabix -b 2 -e 2 -s 1 SAMPLE.gzThis generates two files under “tp/ultra/” directory:
- SAMPLE.gz
- SAMPLE.gz.tbi
Link the file to ProteinPaint by URL for display:
In the URL parameter, set correct genome build version, initial display position, and name and path to the SAMPLE.gz file.
- File path should start from but do not contain “tp/”.
- Name and path is joined by comma.