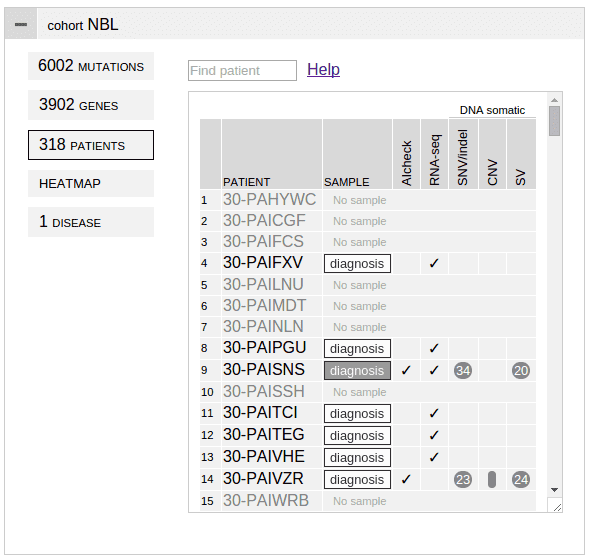
A nicely-organized study. The study is expressed as a JSON file, which contains one single object. To debug JSON, try http://jsonlint.com/.
Example study:
{
"genome":"hg19",
"name":"example study",
"mutationset":[{
"snvindel":"path/to/file.txt",
"name":"DNA mutation"
}],
"show_genetable":1
}Study attributes are listed below, they are case-sensitive.
genome:""
Name of the reference genome, e.g. hg19 or hg38
name:""
The name of this study/cohort.
Assay, tracks, and genome browser
Assays, tracks, and genome browser
"assays":[]
Declares all assay names. Example:
{
"assays":["H3K4me3","RNA-seq","Splice junctions"],
"H3K4me3":{
... ...
},
"RNA-seq":{
... ...
},
"Splice junctions":{
... ...
}
}-
Assay names can be arbitrary words e.g.
- “H3K4me3”
- “RNA-seq”
- “Splice junctions”
- Do not use reserved words such as “assay” or “genome”
- Assay names are case-sensitive
- Each name should appear as an attribute in the study.
-
Each assay is a set of tracks, organized by “individual” → “sample type” → track list
- There can be one or more “sample type” for an “individual”
- In case this is not applicable e.g. for cell lines, use cell line name for both individual and sample type (the hierarchy is hardcoded…)
-
For each sample type, there can be one or multiple tracks associated with it
- If one track, the value is an object
- If multiple tracks, the value is an array of track objects
-
Tracks in one assay should be the same type. Supported types:
- bigWig
- Splice junction
- VCF
- Allelic imbalance (todo)
- Assay tracks can be presented in a sample-by-assay matrix, so called “facet table”. The facet table can be generated by default, and can be customized. See section on facet table.
bigWig tracks
Declare an assay type for the bigWig track, e.g.:
"RNA-seq":{
"config": {
"type": "bigwig",
"pcolor": "red",
"height": 70,
"scale":{ "max":500, "min":0 }
},
"patient1": {
"sampletype1": {
"file": "path/to/RNA-patient1-sampletype1.bw",
"pcolor":"green"
},
"sampletype2": [
... a list of tracks ...
],
.... more sample types
},
... more tracks
},-
config : { }config.typevalue must be “bigwig”- The rest of config contains type-specific configurations that will be applied to every track in this assay. See below link for all available options.
-
[ Patient name ]
- Name of the patient/individual, value is a hash of types of samples from this patient
- Note that the 2-level patient-sampletype arrangement is hard-coded and applies to any type of assay.
-
[ Sample type ]
-
Value could be an object or an array of objects
- Each object is a track
- The track object should contain “file” or “url” to point to the source of the track file
- The track object can contain any configurations and will override the ones in “config”.
-
Read bigWig track configuration options.
Splice junction tracks
Declare an assay type for the junction tracks, e.g.:
"junction":{
"config": {
"type": "junction",
"categories":{
"known":{"color":"#9C9C9C", "label":"Known"},
"novel":{"color":"#cc0000", "label":"Novel"}
}
},
"patient1": {
"sampletype1": {
"file": "path/to/RNA-patient1-sampletype1.gz"
},
.... more sample types
},
... more tracks
},Config.typevalue is “junction”- Config.categories lists types of junctions and their rendering colors
Read junction track configuration options.
VCF tracks
Declare an assay type for the VCF tracks, e.g.:
"VCF":{
"config": {
"type": "vcf"
},
"patient1": {
"sampletype1": {"file": "path/to/patient1-sampletype1.vcf.gz"},
.... more sample types
},
... more tracks
},Additional requirements on the VCF format in ProteinPaint
Assay of AI (allelic imbalance) tracks
To Do
Assay of positional annotation tracks
To Do
In JSON-BED format
E.g. called peaks …
Facet tables
To define what samples and assays are to be used in making a facet table, use the “trackfacets” attribute.
The value is an array, where each element defines one facet table. This way it allows multiple facet tables to be defined.
{
"trackfacets":[
{
"name": "name of this table",
"samples": [ "sample1", "sample2", ],
"assays": [ "assay1", "assay2" ],
"nosortassay":1,
"nosortsample":1
},
... additional facet tables ...
]
}"browserview":{}
Will launch a browser and show some tracks over the given position in the genome.
"browserview":{
"position":{ "chr":"chr10", "start":8081012, "stop":8103311 },
"nativetracks":"refgene,repeatmasker",
"assays":{
"my_rnaseq_assay":1,
"my_splicejunction_assay":{
"combined":1
}
},
"defaultassaytracks":[
{ "assay":"assayname1", "level1":"patientname" },
{ ... another track finder ... }
],
"tracks":[
{ ... track 1 ... },
{ ... track 2 ... }
]
}-
browserview.position- Optional. If not provided, will use default position of the genome.
-
Value can be string or object:
- { chr:”…“, start:…, stop:…}
- “chr1:12345-67890”
- Coordinates are 0-based
-
browserview.nativetracks- Specify what “native” tracks to be loaded by default, usually the gene track. Check with your ProteinPaint server to find out what tracks are available “natively” for that genome.
-
Value can be:
- A string of track name
- A string of multiple track names, joined by comma, no space
- An array of track names
- Track names should match with what’s specified for the tracks of that genome, but case-insensitive
- TODO: allow customizations
-
browserview.assays : { }- Optional. To show all tracks from specified assays.
- If you don’t want to show all tracks of an assay, use “assaytracks” attributes instead (see below)
- Value is an object. Keys are assay names, and the name must exist in the “assays” array.
-
Value to each assay can vary. In above example:
- By using value “1”, all tracks from “rna_seq” will be displayed as separate tracks.
-
By using value {”combined“:1}, all tracks from this assay type will be aggregated into one track. This only applies to these track types:
- Junction
- VCF
-
browserview.defaultassaytracks : [ {} ]- Optional. To select a set of tracks from the assays defined in this study, and show them by default.
-
Each element is a “track finder”, with these attributes to pinpoint the track:
-
”assay”:""
- The assay name, required.
-
”level1”:""
- Patient name, but here it’s a generic name. Required.
-
”level2”:""
- Optional, the sample type. If specified, will use track from only this sample type of the patient as specified by “level1”. Else, it will use all tracks from this patient.
-
-
browserview.tracks : [ ]- Optional. To provide a list of custom tracks.
- Custom tracks defined as JSON objects, see JSON format of tracks.
- This allows adding tracks in freedom, directly encoded, rather than systematic way of “assays”. Such tracks are not associated with any assay type and won’t show up in facet table.
Mutation data in tabular format
mutationset:[]
Lists all mutation datasets for this cohort, and the mutation data files associated for each set.
"mutationset":[
{
"name":"DNA somatic",
"snvindel":"datadir/snvindel_dna",
"sv":"datadir/sv",
"cnv":"datadir/cnv"
},
{
"name":"RNA-seq",
"snvindel":"datadir/snvindel_rna",
"fusion":"datadir/fusion"
}
],This example provides two sets of mutation for a cohort from both DNA and RNA, as indicated in the name. The name can be arbitrary string. In each dataset, different types of mutations are provided in the form of separate text files.
The “datadir” from above example is relative to the ROOT directory.
Mutation data type keys, each should point to a server-hosted text file.
-
snvindel
- SNV/indel data, tabular text format
- sv
-
fusion
- Structural variation OR fusion, they are in the same tabular text format
- Note that “sv” is used in “DNA”, and “fusion” is used in “RNA-seq”
-
svfusion
- JSON format data for sv/fusion/ITD/deletion/truncation, format is the same as the JSON file made by fusion editor export.
-
cnv
- Copy number change, tabular text format
-
deletion
- Intragenic deletion, tabular text format
-
truncation
- Gene truncation, tabular text format
-
Variantgeneassoc
- No longer supported!!
- For describing genes associated with particular variants
- Per-sample based: each line is one variant from one sample.
- This will trigger a browser view for exploring variant and genes together.
-
7 columns:
i. Chromosome
ii. Position
iii. Reference_allele
iv. Mutant_allele
v. Patient
vi. Sampletype
vii. geneset
-
The “geneset” is a JSON array, representing associated genes from each sample, example:
i. [{“name”:“TAL1”,“isoform”:“NM003189”,“position”:“chr1:47681961-47698007”,“score1”:0.543,“score_2”:0.543}, {… second gene …} ]
dbexpression:{}
It provides a gene-level expression values over a group of samples from a database table on the server, and show it when the gene is shown in the protein-view.
“Dbexpression” should work with “mutationset”.
"dbexpression":{
"dbfile":"path/to/data.db",
"tablename":"[name of the table]",
"keyname":"[name of the table field to query against, e.g. gene]",
"tidy":"function(rows){rows.forEach(function(row){ ... })}",
"config":{
"name":"RNA-seq",
"sampletype":"sample",
"datatype":"FPKM",
"ongene":true,
"hlcolor":"#f53d00",
"hlcolor2":"#FFBEA8",
"attrlst":[{"k":"patient"},{"k":"sampletype"}],
"cohort":{
"levels":[
{"k":"disease","label":"Disease"}
]
}
}
},-
dbexpression.dbfile
- Path to the SQLite database file. Must be relative to the
directory.
- Path to the SQLite database file. Must be relative to the
-
dbexpression.tablename
- Name of the database table to query against with
-
dbexpression.tidy
- Provides a Javascript function in the form of a string, argument is an array of all records fetched from the database, the function will apply certain tidying operation on each of the record
-
dbexpression.attrlst
Note that the db table can contain data for samples that are not from this cohort, and they will still be displayed in the expression data panel.
Gene-sample heatmap
heatmapJSON:{}
An JSON object that configures the heatmap layout. Refer to heatmap configuration tutorial.
hardcodemap:[]
Provides one or more “hard-coded heatmap”.
"hardcodemap":[
{
"file":"path/to/file.txt",
"rowh":18,
"rowspace":1,
"colw":10,
"colspace":1,
"metadata":{
"key1":{
"v1":{"label":"what","color":"red"},
"v2":{"label":"what","color":"blue"}
},
"key2":{
"V3":{"label":"what","color":"red"},
"V4":"{"label":"what","color":"red"}
}
}
],File format:
- Columns are samples
- Rows are items (e.g. genes, or other attributes describing samples)
-
First line is header
- First column is metadata keys, the value must be found as one of the keys in hardcodemap.metadata
- Second column is row name
- Cell value is one of the values of each key (of that row). Join multiple values by semicolon.
Metadata annotation
(under development)
"patientannotation":{}
An object to define metadata annotation at patient level.
This should also be used when there is only information on samples but not patients (in which case sample name/id should be expressed at “patient” level).
patientannotation.annotation provides actual annotation of each patient/sample.
patientannotation.metadata provides the list of metadata terms, and attributes associated with the term.
"patientannotation":{
"annotation":{
"patient1":{
"age":"child",
"sex":"m"
},
... more patients ...
},
"metadata":[
{
"key":"age",
"label":"Age at diagnosis",
"values":[
{
"key":"child",
"label":"Child (<10yrs)",
"color":"red"
},
{
"key":"adult",
"label":"Adult (>18yrs)",
"color":"blue"
}
]
},
{
"key":"sex",
"label":"Sex",
"values":[
{
"key":"m",
"label":"Male",
"color":"red"
},
{
"key":"f",
"label":"Female",
"color":"blue"
}
]
},
... more terms ...
]
}Such metadata annotation can be applied to:
- Expression - PCA plot
- Splice junction track (TODO)
"annotations":{}
An object to define metadata annotation by input file.
.idkey "STRING"
The column value, typically “sample” or “samplename”, to use for collecting annotations into an object
.files [ARRAY]
An array of string filepaths to use as metadata source data
inputFormat "STRING"
Indicates the format for parsing the source file, defaults to “metadataTsv”
Expression - PCA plot
"e2pca":[]
A list of plots to be made, each defines an “expression - PCA” plot.
"e2pca": [
{
"name": "optional name of this plot",
"vectorfile": "path/to/vectorfile.txt",
"dbfile": "path/to/data.db",
"colorscale":{
"from":"blue",
"to":"red"
}
},
... one more plot ...
],Other controls
sankey:{}
An object of possible configurations to the disease-gene sankey diagram.
Use sankey.genes:[] to define a set of genes to be used in the diagram:
"sankey":{
"genes":[
{"name":"ERG","color":"red"},
{"name":"ZEB2","color":"red"},
{"name":"MYC","color":"red"},
{"name":"MYCBP2","color":"red"}, ....
]
},Use sankey.geneset:[] to define gene sets to be used in the diagram, all genes in a set share the same color, legend will be rendered:
"sankey":{
"geneset":[
{"name":"gene set 1",
"color":"red",
"genes":["ERG","ZEB2"]
},
{"name":"gene set 2","color":"blue","genes":[ ...... ]}
]
},diseasecolor:{}
An object to define rendering color for different diseases. Applies to sankey diagram and gene network chart.
genenetwork:{}
To provide one or more gene network diagrams.
"genenetwork":{
"list":[
{ a JSON object representing a network },
{ one more network }
]
}Each network is defined as a JSON object, see example.
no_defaultgeneset:1
Will not apply the genome-equipped gene set to generate the heatmap view, but rather, to use highly recurrent genes based on the data:
- Max 20 genes, all in one group.
show_genetable:1
Will show the gene table by default. Only applicable when “mutationset” is used. Value is “1” for true.
show_sampletable:1
Will show the sample table by default. Value is “1” for true.
Samples can come from either “mutationset”, or assays.
disable_sampletable:1
Will hide the sample table.
show_heatmap:1
Will show the heatmap by default. Value is “1” for true.
show_browser:1
Will show the genome browser by default. Value is “1” for true.
show_hardcodemap:1
Will show the hard-coded heatmap by default.
"show_e2pca":1
Will show the expression - PCA plots by default.
disable_genenetwork:1
Will hide the gene network function.
hide_addnewfile:1
Will hide the “+NEW FILE” tab.
hide_navigation:1
Will hide all navigation buttons on the left.
individual_label_name:"patient"
Will replace the default “individual” with the given word in the ”# individuals” tab on left of the study view.
Obsolete contents
STOP HERE.
warning
The rest of these contents are obsoletse. We’re busy porting them to the new ProteinPaint.
Attribute: aicheck (moved to "assays")
This is the “aicheck” track.
This is the integrative display of allelic frequency imbalance of variants or genetic markers, as well as DNA sequencing coverage over these markers. Adapted from the “aicheck” figure invented by Xiaotu Ma.
Note that this is designed for tumor-normal matched paired samples only.
"aicheck": {
"20-PABLDZ": {
"sampletypes": [
"relapse",
"diagnosis"
],
"relapse": {
"file2": "aml/ai_check_SJBALL013790_R1_G1.txt.gz",
"readdepthcutoff": 126
},
"diagnosis": {
"file": "aml/fullMaf_TARGET-20-PABLDZ-09A-02D_NormalVsPrimary.maf.txt.gz",
"readdepthcutoff": 126
}
},
"patient2":{ ... },
... more patients ...
},The “samplekeys” is an array of sample type. This is a relic of initial design and will be abandoned. By then it will be ignored by ProteinPaint.
There are two file formats for aicheck, identified by “file” and “file2”. “file2” format comes right off the CompBio pipeline (generated by one of Xiaotu’s script).
“file” columns
- chromosome name, must be “chr1” but not “1”
- coordinate, 1-based
- MinD: mutant allele read count in tumor DNA
- TinD: total read count in tumor DNA
- MinN: mutant allele read count in normal DNA
- TinN: total read count in normal DNA
“file2” columns
- chromosome name, must be “chr1” but not “1”
- coordinate, 1-based
- SNP, value seems always is “SNP”
- rsNumber: dbSNP name
- TinD: total read count in tumor DNA
- dMAF: mutant allele frequency in tumor DNA
- TinN: total read count in normal DNA
- nMAF: mutant allele frequency in normal DNA
The “readdepthcutoff” value defines the Y-axis max value of read depth track for both tumor and normal. It is pre-calculated by following Xiaotu’s method below. User is free to use any other method.
######## to calculate trimmed media, in 1% to 99% range
trimed.median<-function(xx) {
cutt<-0
dnn<-quantile(xx,0.01)
upp<-quantile(xx,0.99)
if(sum(xx>dnn & xx<upp)>2) {
use<-(xx>dnn & xx<upp)
cutt<-quantile(xx[use],0.98)
}
return(as.numeric(cutt))
}
#Tumor
medd.D<-trimed.median(dt0[,"TinD"])*1.5
#Normal
medd.G<-trimed.median(dt0[,"TinN"])*1.5
if(medd.D<1) medd.D<-1
if(medd.G<1) medd.G<-1
#Maximum
medd<-max(medd.D,medd.G)Broadly, there are two types of tracks that ProteinPaint can read: numerical, and positional. Bedgraph and bigWig are numerical track formats, the generic bed is the positional format.
In the cohort definition, you have to first declare what the tracks are about, then the type/format of the track. The first layer is assay type, like RNA-Seq, or it can be anything with a name. We will refer to it as “assay type”.
Assay types are predefined, allowed values are (case-sensitive):
- rnaseq
Each assay type must be declared as a hash, with patient names as keys. Patients from different assay types may or may not be the same.
There is always a reserved key called “config”, providing configurations applicable to all tracks of this assay type. Each track can have its own configurations in order to be different from the “global” attributes in config.
Numerical track Y-scale configuration
-
percentile
- Value is positive integer from 1 to 99
- If provided, the nth percentile value in the view range will be used to set Y-axis. This is useful for preventing large outlier values from skewing the display.
- When there are both positive and negative values in the view range, the percentile calculation will be applied separately to positive and negative values for calculating max and min values.
-
min, max
- When valid values are provided for both, will set fixed scale.
-
autoscale
- Apply auto-scale, will override percentile or min/max settings.
File or URL?
-
url
- HTTP or HTTPS, such as http://datahub.stjude.org/datasets/MBSE2016/hg19/tracks/1M1H3K27Actreatafterfitingall.bw
- The hosting server must support byte-range request
-
file
- A partial path to a file, such as “path/to/track.gz”, relative to the Tumor Portrait root directory
-
ProteinPaint will reject:
- absolute path (e.g. /etc/passwd)
- back-to-previous-level (e.g. ../../etc/passwd)
Track: bedGraph
bedGraph track example for RNA-Seq
```javascript
"rnaseq":{
"config":{
"name":"RNA-seq coverage",
"tktype":"bedgraph",
"pcolor":"#006600",
"pcolor2":"#CC00B8",
"percentile":95
},
"patient1": {
"diagnosis":{
"file":"aml/SJAML040582_R1.gz"
},
"relapse":{
"file":"somewhere/xxxxx.gz"
}
},
"patient2": { ... },
... more patients ...
},
```Track: bigWig
bigWig example for RNA-Seq
```javascript
"rnaseq":{
"config":{
"name":"RNA-seq coverage",
"tktype":"bigwig",
"pcolor":"#006600",
"pcolor2":"#CC00B8",
"percentile":95
},
"patient1": {
"sampletype":{
"file":"aml/SJAML040582_R1.bw"
}
},
... more ...
},
```Track: numeric2 (a pair of numerical tracks)
Overlaying of two numerical tracks, noted by “track1” and “track2”.
-
Track 1
- Displayed on foreground, axis on left
-
Track 2
- Displayed on background, axis on right
rnaseq coverage-FPKM overlay example
```javascript
"rnaseq":{
"config":{
"name":"RNA-seq",
"tktype":"numeric2",
"track1":{
"name":"coverage",
"rangelimit":10000000,
"pcolor":"#006600",
"pcolor2":"#CC00B8",
"percentile":95
},
"track2":{
"pcolor":"#FF9900",
"pcolor2":"#CC7A00",
"name":"FPKM",
"autoscale":"on"
}
},
"patient1": {
"diagnosis":{
"file":"aml/SJAML040582_R1.gz",
"file2":"aml/10-PAPAIZ-diagnosis.gz"
}
},
"patient2": { ... },
... more patients ...
},
```Track: junction (RNA-Seq junction reads)
junction example, along with a “browserview” trigger for displaying
```javascript
"junction": {
"config":{
"type2color":{
"known":"#9C9C9C",
"novel":"#cc0000"
}
},
"30-PAIFXV": {
"diagnosis": {
"file": "junction/targetNBL/30-PAIFXV-diagnosis-SJNBL017066_D1.gz"
}
},
"30-PAIPGU": {
"diagnosis": {
"file": "junction/targetNBL/30-PAIPGU-diagnosis-SJNBL017070_D1.gz"
}
},
...
},
"browserview":{
"position":{ "chr":"chr12","start":25357723,"stop":25403865 },
"assays":{
"junction":{
"sum_view":{
"type":"junction",
"name":"NBL"
}
}
}
}
```Each junction track file contains splicing junctions identified in ONE SAMPLE only. Primarily this is converted from the RNApeg output (http://hc-wiki.stjude.org/display/compbio/How+to+count+novel+or+reference+junction+reads+in+an+RNA-Seq+BAM+using+RNApeg).
To convert an RNApeg output file into a track file to be displayed on ProteinPaint, run:
node utils/rnapegjunction2tabix.js sample.RNApeg.output sample.junctionThe converted file has 5 columns:
- Chromosome name, e.g. “chr1”
- Start, 1-based position of the last exon nucleotide
- Stop, 1-based position of the first exon nucleotide
- Number of (high-quality) junction reads
-
Type of junction
a. RNApeg outputs two types of junction (known/novel). For displaying, arbitrary types can be used. Any used types should be stated in the config.jtype2color so they can be distinguished by color
Track: VCF
VCF examples, along with “browserview” trigger for displaying
```javascript
"vcf_cohort":{
"file":"vcf/scd_143_join.vcf.gz",
"name":"Joint"
},
"vcf":{
"SJSCD040763":{
"G1":{
"file":"vcf/SJSCD040763_G1.vcf.gz"
}
},
"SJSCD040764":{"G1":{"file":"vcf/SJSCD040764_G1.vcf.gz"}},
...
},
"browserview":{
"position":{"chr":"chr6","start":135179289,"stop":135183700},
"assays":{
"vcf_cohort":1,
"vcf":{
"sum_view":{
"type":"vcf",
"name":"Individual"
}
}
}
}
```“Vcf_cohort” specifies one single VCF file.
“Vcf” specifies the VCF file from each sample.
Track: vafs1 (variant allele fraction of a single sample)
vafs1 example
"vafs1":{
"person1":{
"sampletype1":{
"DNA":{"file":"variantgene/SJALL040467_D1.combine.WGS.goodmarkers.txt.gz"},
"RNA":{"file":"variantgene/SJALL040467_D1.combine.RNAseq.goodmarkers.txt.gz"}
},
"sampletype2":{
"DNA":{"file":"variantgene/SJALL040467_R1.combine.WGS.goodmarkers.txt.gz"},
"RNA":{"file":"variantgene/SJALL040467_R1.combine.RNAseq.goodmarkers.txt.gz"}
}
},
... next person ...
}Track file has 6 columns:
- Chromosome
- Position (1-based)
- Reference allele
- alternative allele
- Total reads
- Variant allele fraction
Note that for one sample type, there can be multiple types of tracks (DNA/RNA in above example). The type names can be arbitrary.
Track: BAM
In progress …