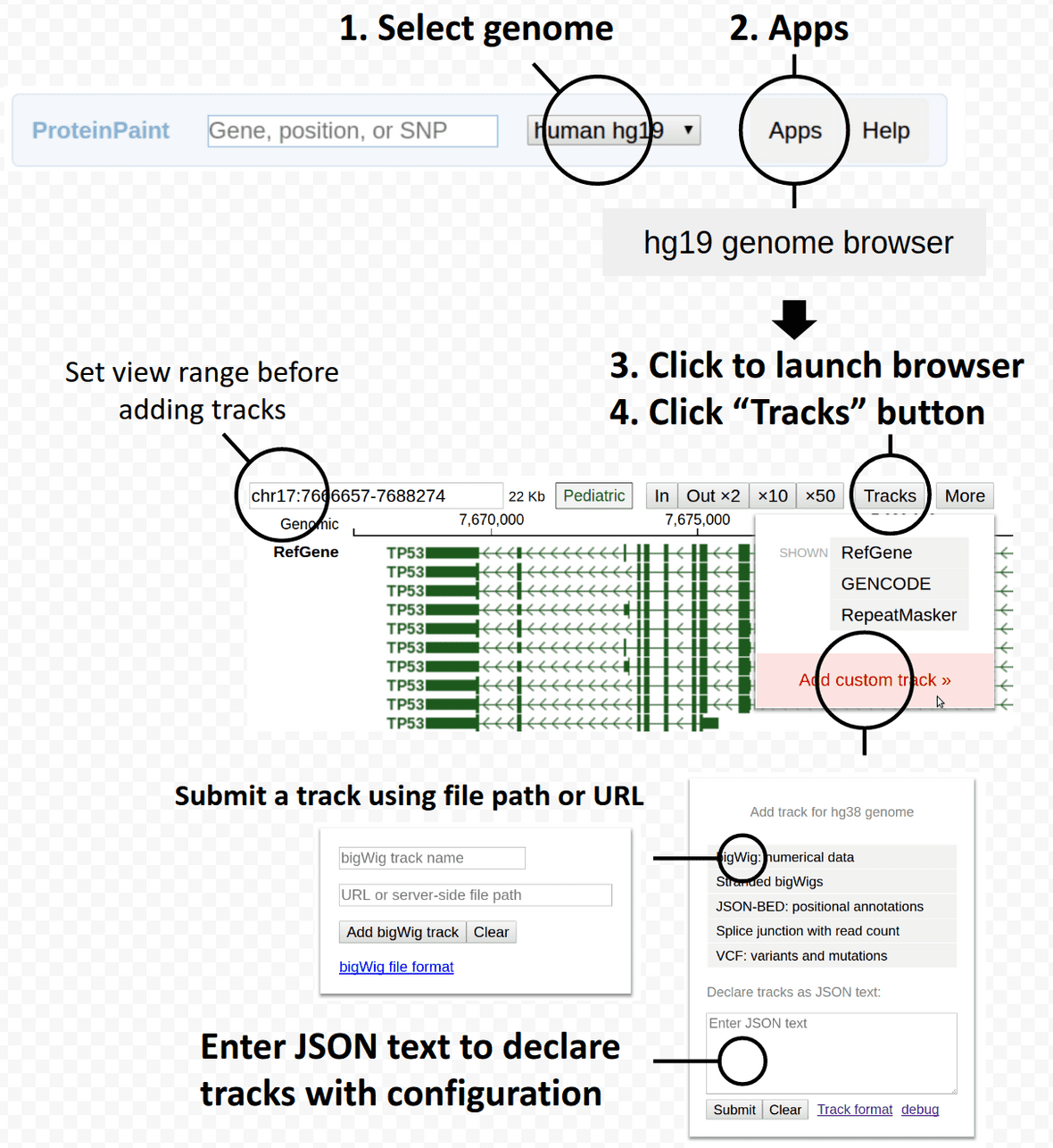
Using custom tracks in ProteinPaint
- Tracks are defined as JSON objects.
- Submit tracks by launching a genome browser, see below:
- All text values are case-sensitive.
Use these color names
- Common names such as red, green: https://en.wikipedia.org/wiki/Web_colors
-
FF0000
- rgb(255,0,0)
- rgba(255,0,0,.5)
Example
Go to https://proteinpaint.stjude.org, launch hg19 genome browser and paste in following JSON text to add two tracks:
[
{"type":"bigwig","file":"hg19/hg19.100way.phastCons.bw","name":"UCSC phastCons 100ways","dotplotfactor":20,"height":100},
{"type":"bedj","file":"anno/refGene.hg19.gz","name":"RefSeq genes","translatecoding":1,"color":"#417D4C","stackheight":20}
]View or debug JSON with https://jsonlint.com/
The JSON track objects can be used with embedding API.
Gallery
https://proteinpaint.stjude.org/examples/
Attributes applicable to all track types
Example:
{
"name":"name of the track",
"type":<typecode>,
"url":"http://domain/file.gz",
"indexURL":"http://domain/path/file.gz.tbi",
"file":"path/to/file.gz", // use this when not using URL
"toppad":5,
"bottompad":5
}-
”name”: STR
- A string as track name
-
”type“:
-
Typecode of the track. Allowed values are:
- bigwig
- bigwigstranded
- bedj
- profilegenevalue
- junction
- vcf
- bampile
-
- ”file”: STR
- ”url”: STR
-
”indexURL”: STR
- Either “file” or “url” should be provided, but not both. When using “file”, provide the relative path to the track file starting from
directory as is configured on the ProteinPaint server. - When using URL for tabix-indexed files, by default it requires the index file to share URL with the .gz file. When it’s not sharing the URL, the attribute “indexURL” must be used to provide the URL of the index file.
- Either “file” or “url” should be provided, but not both. When using “file”, provide the relative path to the track file starting from
-
”toppad”: INT
- Number of pixels as the padding space on the top, default: 5
-
”bottompad”: INT
- Number of pixels as the padding space at the bottom, default: 5
-
”hidden”: 1
- If set, the track will be hidden by default and can be found in the track menu (by clicking the “Tracks” button)
Track: bigWig for numerical data
Example:
{
"name": "track name",
"type": "bigwig",
"file": "targetRNAcoverage/SJNBL017066_D1.bw",
"scale": {
"min": 0,
"max": 100
},
"height": 100
}Live example: https://proteinpaint.stjude.org/examples/bigwig.html
bigWig track attributes:
-
scale : { }
- min
-
max
- Set a fixed scale range of the Y axis
-
percentile
- Value is integer from 1 to 99, representing a percentile of all the data in the view range
- Overrides min/max
-
auto
- Value is simply “1” for “on”.
- Set automatic scale, will override all other settings in “scale”
-
height
- Bar plot height in number of pixels.
- If height is below 10, the track will be rendered as heatmap.
-
pcolor
- Bar color of the positive values
-
pcolor2
- Rendering color for data points above Y axis maximum value.
-
ncolor
- Bar color of negative values
-
ncolor2
- Rendering color for data points below Y axis minimum value
-
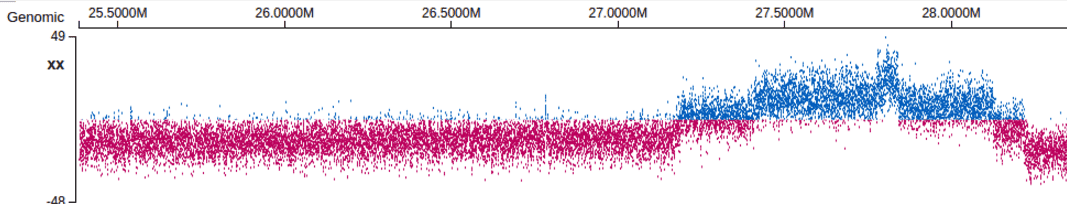
dotplotfactor
- Value is positive integer e.g. 5 or 10. When applied, will request 5 or 10 times more data points from a bigWig track and plot each point as a dot, rather than bars. A use case is checking (large-scale) CNV from DNA sequencing coverage track
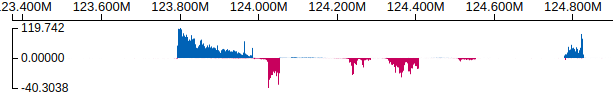
Track: a pair of bigWig for data from forward and reverse strands
Example: https://proteinpaint.stjude.org/examples/bigwig.stranded.html
For showing stranded RNA-Seq coverage data as a pair of bigWig tracks, with forward strand on top and reverse strand on bottom.
{
"name": "stranded RNA-seq coverage",
"type": "bigwigstranded",
"strand1":{
"file": "path/to/sample.forwardstrand.bw",
"scale": {
"min": 0,
"max": 100
},
"height": 50
},
"strand2":{
"file": "path/to/sample.reversestrand.bw",
"scale": {
"max": 0,
"min": -100
},
"height": 50,
"normalize": { "dividefactor": -1 }
}
}-
strand1 : {}
- The bigWig track of the data from forward strand
- For read-coverage data, the values in the forward-strand bigWig file should be positive
-
strand2 : {}
- The bigWig track of the data from reverse strand
- Both strands follow the bigWig track definition.
Note: for stranded bigwig files using all positive values for both strands (e.g. sequencing read coverage), a “normalization value” of -1 should be applied to the reverse strand, so the bars will point down.
If the reverse strand bigwig track has been prepared to have negative values, then no need to apply the -1 normalizing factor.
Track: JSON-BED
Example: https://proteinpaint.stjude.org/examples/bedj.html
The JSON-BED track for annotating genomic features, e.g. genes.
Full specification of the JSON-BED track format.
Track: genomic profile combined with gene expression value (PGV)
Example: https://proteinpaint.stjude.org/examples/pgv.html
Track: splice junction
Example: https://proteinpaint.stjude.org/examples/junction.html
{
"type": "junction",
"name": "sample junction",
"file": "junction/targetALL/10-PANYGB-diagnosis-SJCOGALL010859_D2.gz",
"categories": {
"known": {
"label": "Known",
"color": "#9c9c9c"
},
"novel": {
"label": "Novel",
"color": "#cc0000"
}
}
}“categories” specifies the rendering color for types of junctions.
Read the splice junction file format.
Multiple junction tracks can be aggregated to show in one track, via the .tracks[ ] attribute:
{
"type":"junction",
"name":"sample junction",
"tracks":[
{
"sample":"sample1",
"file":"path/to/sample1.gz"
},
{
"sample":"sample2",
"file":"path/to/sample2.gz"
},
... more samples ...
],
"categories":{ ... }
}In the .tracks[ ], add one object for each member track.
When combining multiple junction tracks, each track can contain one or multiple samples.
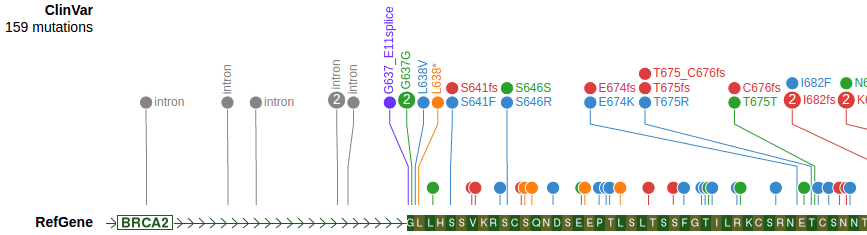
Track: VCF
Example: https://proteinpaint.stjude.org/clinvar.html
{
"name": "track name",
"type": "vcf",
"file": "path/to/vcffile.gz",
"url": "use url if the vcf file is hosted on a web server"
}- SNV and indel variants only, other types are not yet supported, e.g. CNV and SV.
- A VCF file can contain one or multiple samples.
ProteinPaint requires VCF format specification version 4.2
Please refer to this document on following topics about the VCF track
- Preparing and hosting a VCF file
- Encoding functional annotation for coding-region mutations
- Combining multiple VCF files into one track
- Highlighting and filtering variants by values of an INFO field
- Features available for multi-sample VCF track a. Population frequency filter b. Sample annotation c. VAF-coverage plot
Track: bampile
Bampile track can be used to examine the very rare alleles from a high-depth (capture-based) DNA sequencing experiment. Example of bampile track:
{
"name": "track name",
"type": "vcf",
"file": "path/to/vcffile.gz",
"url": "use url if the vcf file is hosted on a web server"
}-
fineheight
- Height of the bar plot at bottom showing low-frequency alleles, the Y scale uses a cutoff value as defined by “fineymax”
-
allheight
- Height of the bar plot at top showing frequency of all alleles with automatic scale for coverage
-
midpad
- Padding distance between top and bottom bar plots
-
fineymax
- Y scale used by the bottom bar plot
-
usegrade
- Name of the grade to use
Track: aicheck
Example: https://proteinpaint.stjude.org/examples/aicheck.html
Read the aicheck track tutorial.
Track: GenomePaint
Example: https://proteinpaint.stjude.org/examples/svcnv.html
This launches custom MDS track (aka GenomePaint). To access official tracks, see embedding API.
{
"name": "track name",
"type": "mdssvcnv",
"file": "path/to/svcnv.gz",
"checkexpressionrank":{
"file":"hg38/tcga-gdc/SKCM/TCGA_SKCM.fpkm.gz"
}
"checkvcf":{
"file":"hg38/tcga-gdc/SKCM/TCGA_SKCM.vcf.gz"
}
}Read the GenomePaint tutorial.
You can replace “file” with “url” in above 3 places.
Following attributes can be applied in the track object as detailed in the Embedding API.
- singlesample:{}
- isfull:true / isdense:true
- sampleAttribute:{}
- vcf:{}
- hide_cnvgain:true
- hide_cnvloss:true
- cnv:{}
- sampleset:[]
A derivative of the “mdssvcnv” track is the multi-sample ASE track.
Example: https://proteinpaint.stjude.org/examples/ase.html
{
type:'mdssvcnv',
name:'Multi-sample ASE analysis',
checkvcf:{
file:'hg19/TARGET/DNA/test/oct3/sorted.vcf.gz',
},
checkrnabam:{
samples:{
SJALL015260_D1: {
file:'hg19/TARGET/RNAbam/SJALL015260_D1.bam',
totalreads: 83388794,
},
SJALL015643_D1: {
file:'hg19/TARGET/RNAbam/SJALL015643_D1.bam',
totalreads: 103477133,
},
... more BAM files ...
},
}Track: ASE, single-sample
Example: https://proteinpaint.stjude.org/examples/ase.single.html
This carries on-the-fly ASE analysis for a single sample.
This track can be built standalone, or spawned from a multi-sample track (a special mode of mdssvcnv track)
{
type:'ase',
name:'My sample ASE',
samplename:'my_sample_name',
rnabamfile:'path/to/sample.rnaseq.bam',
rnabamtotalreads: 103477133,
vcffile:'path/to/SJALL015643_D1.gz',
},