Loading TCGA MAF file
The MAF format is extensively used by TCGA and others to present cancer genomic mutations. The MAF format is specified here: https://wiki.nci.nih.gov/display/TCGA/Mutation+Annotation+Format+(MAF)+Specification. Here we describe steps to download MAF files from the TCGA Data Portal and display on ProteinPaint.
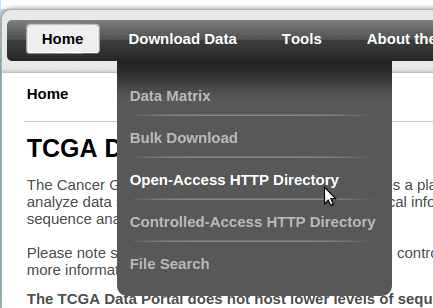
Visit the TCGA data portal at https://tcga-data.nci.nih.gov/tcga/ and go to the “Open-Access HTTP Directory” from the menu:
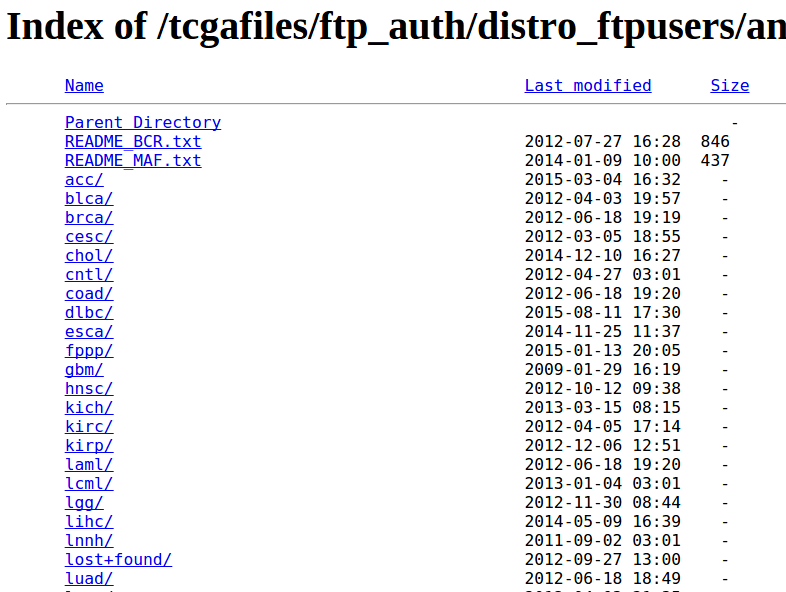
In the HTTP directory, click on any of the disease folder to look for mutations stored in MAF files:
You may need to go down several folders to find the MAF files. For skin cancer, the path of folders to find MAF files is:
skcm/
> gsc/
> broad.mit.edu/
> illuminaga_dnaseq/
> mutations/
> broad.mit.edu_SKCM.IlluminaGA_DNASeq.Level_2.0.1.0/As an example, from this folder we download a MAF file named “PRTCGASKCMPAIRCaptureAllPairs_QCPASS.aggregated.capture.tcga.uuid.somatic.maf”.
This MAF file can be submitted to ProteinPaint. Due to the relatively large size of this MAF file, it can be helpful to keep only essential columns so as to reduce file size and speed up uploading and subsequent browsing. This can be done either using a shell command, or a spreadsheet software.
Using shell command:
cut -f1,5,6,9,16,40,42,44 PR_TCGA_SKCM_PAIR_Capture_All_Pairs_QCPASS.aggregated.capture.tcga.uuid.somatic.maf > TCGA_SKCM.slim.mafUsing a spreadsheet software (e.g. Microsoft Excel, OpenOffice):
- Import the MAF file into the software by choosing “tab” as delimiter.
- Select columns with following header names: HugoSymbol, Chromosome, Startposition, VariantClassification, TumorSampleBarcode, cDNAChange, ProteinChange, RefseqmRNA_Id. Copy the contents of selected columns.
- Create a new sheet and paste these columns into the new sheet.
- Choose “Save as…” and select “tab-delimited text” format to save this sheet to a text file.
Both methods will generate a new file which is 10% the size of the original MAF file. This new file contains following columns. Note that ProteinPaint does not require any specific order on the columns:
- Hugo_Symbol
- Chromosome
- Start_position
- Variant_Classification
- TumorSampleBarcode
- cDNA_Change
- Protein_Change
- RefseqmRNAId
At the time of writing, ProteinPaint does not support disease subtype and subgroup information, as well as fusion gene data from the uploaded text file. Please check with the latest instructions on ProteinPaint to find out all supported data types and attributes.
Uploading MAF file to ProteinPaint
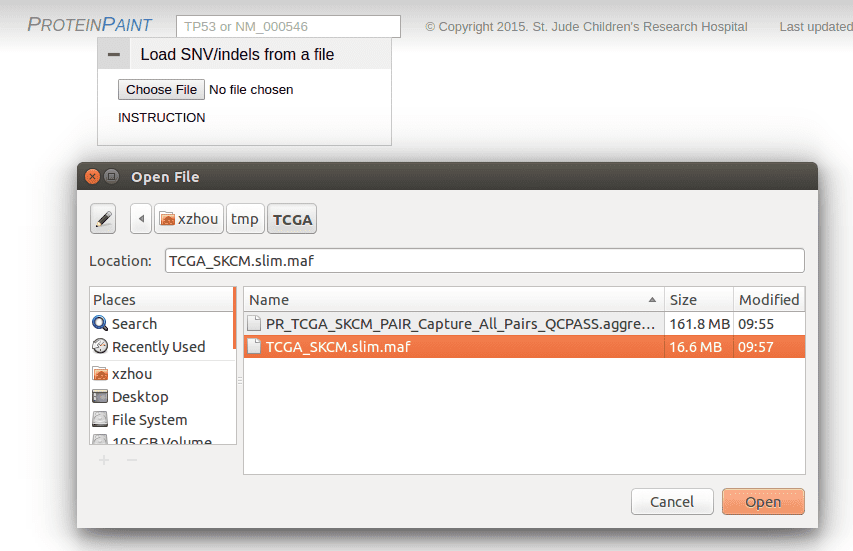
Load the trimmed-down version of the MAF file to ProteinPaint using the file upload panel previously shown:
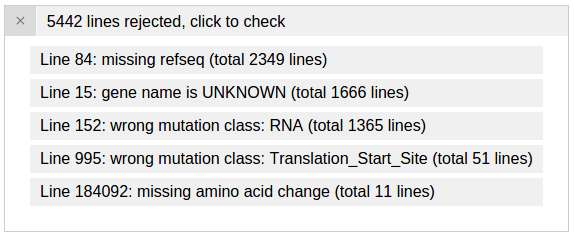
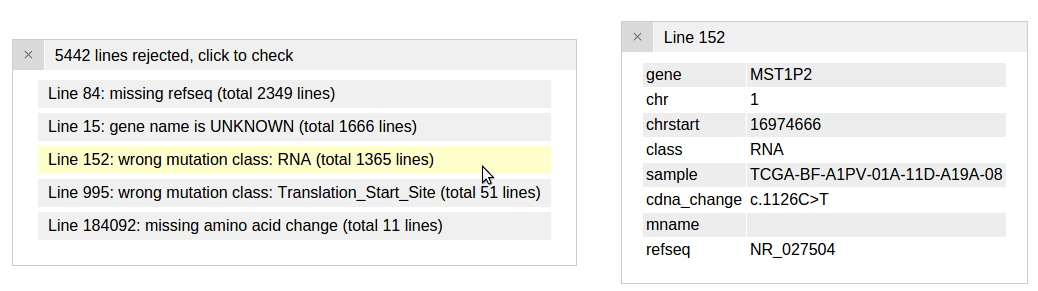
After loading this file, two new panels appear. The first panel shows the 5 reasons that some lines from the uploaded file are rejected by ProteinPaint. The reasons are ranked by the number of lines in which they occur:
Click the first entry to inspect a rejected line for the reason “wrong mutation class: RNA”. This is because that “RNA” describes a mutation in a noncoding gene, which ProteinPaint currently does not support:
Please refer to other tutorials on exploring the data content from an uploaded MAF file.